News
May 2025
Miniconference entitled EXBIO25 (Experimental Biology 2025) took place in our laboratory. Students presented milestones in their bachelor's and diploma theses and were awarded with mini-certificates for their effort and active participation.



24-10-2023
A new research paper published as a result of great collaboration with Dr. Thomas M. Jovin and Dr. Donna A. Jovin from the Max Planck Institute for Interdisciplinary Sciences
https://www.mdpi.com/1422-0067/24/13/10740
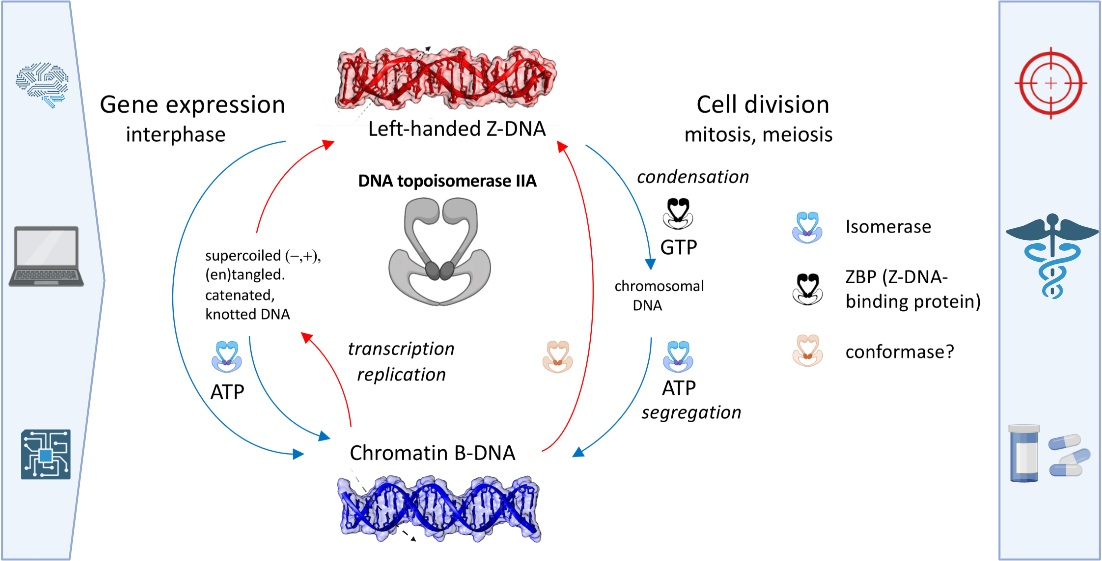
In this research paper, titled "Extensive Bioinformatics Analyses Reveal a Phylogenetically Conserved Winged Helix (WH) Domain (Zτ) of Topoisomerase IIα, Elucidating Its Very High Affinity for Left-Handed Z-DNA and Suggesting Novel Putative Functions," our research group investigates the fascinating novel properties of the Winged Helix (WH) domain, specifically the Zτ variant found in Topoisomerase IIα, which could have an implication for the development of a new class of Topoisomerase II inhibitors/drugs.
17-10-2021
Visit our YouTube channel 🙂
https://www.youtube.com/channel/UCBHnfRaRO2cgPVSqF_swS8A
18-9-2021
Contribute to our new special IJMS issue! 🙂
"Bioinformatics of Unusual DNA and RNA Structures"
https://www.mdpi.com/journal/ijms/special_issues/Unusual_DNA_RNA
- Journal Rank: JCR - Q1 (Biochemistry & Molecular Biology) / CiteScore - Q1 (Inorganic Chemistry)
The deadline for submissions is 31 March 2022

19-7-2021
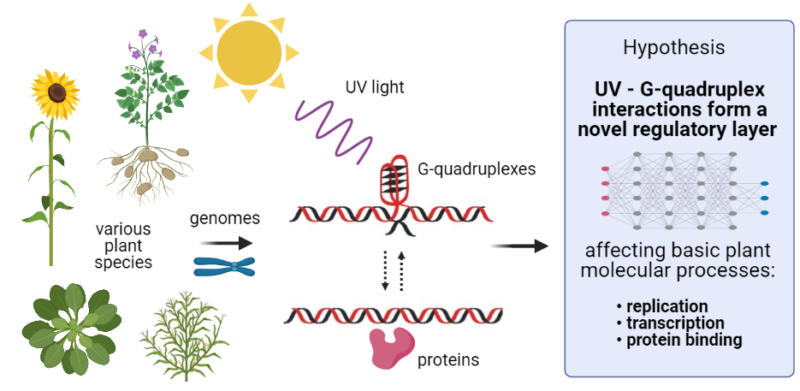
G-Quadruplex in Gene Encoding Large Subunit of Plant RNA Polymerase II: A Billion-Year-Old Story, our current paper just published in the International Journal of Molecular Sciences (impact factor 5.9). Great congrats to the first author Ms. Adriana Volná, and corresponding author Dr. Jiří Červeň! 🙂
full-text freely available here: https://www.mdpi.com/1422-0067/22/14/7381
22-3-2021
We have a new logo for our lab (DNA-Chess on the front page)! Many thanks to our amazing artist, lady M.Sc. Alena Volná 😘
Alena Volná studied special pedagogy at the University of Ostrava, but she has been intensively involved in fine arts since her youth. Recently, she completed certified art courses focused on the so-called "Dry brush" (i.e. drawing with oil paints). She prefers to work with pencils and watercolors. She currently works as a school club leader and art teacher at a private elementary school. As part of leisure activities for children, she also leads a hobby group of artistic and technical activities, where, in addition to drawing, she also devotes herself to ceramics and work with alternative materials (wood, etc.) together with children. She supplemented the last few professional publications of our team (e.g. p53, SARS-CoV-2) with her illustrations.


20-1-2021
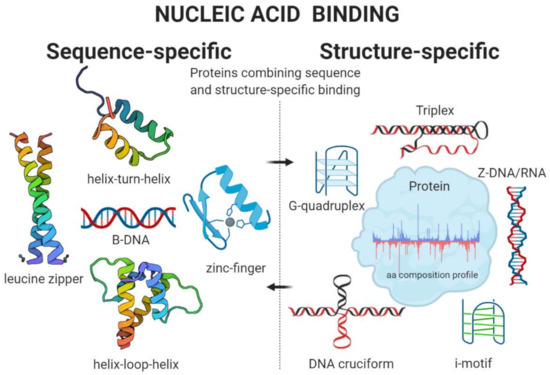
Amino Acid Composition in Various Types of Nucleic Acid-Binding Proteins
Abstract:
Nucleic acid-binding proteins are traditionally divided into two categories: With the ability to bind DNA or RNA. In the light of new knowledge, such categorizing should be overcome because a large proportion of proteins can bind both DNA and RNA. Another even more important features of nucleic acid-binding proteins are so-called sequence or structure specificities. Proteins able to bind nucleic acids in a sequence-specific manner usually contain one or more of the well-defined structural motifs (zinc-fingers, leucine zipper, helix-turn-helix, or helix-loop-helix). In contrast, many proteins do not recognize nucleic acid sequence but rather local DNA or RNA structures (G-quadruplexes, i-motifs, triplexes, cruciforms, left-handed DNA/RNA form, and others). Finally, there are also proteins recognizing both sequence and local structural properties of nucleic acids (e.g., famous tumor suppressor p53). In this mini-review, we aim to summarize current knowledge about the amino acid composition of various types of nucleic acid-binding proteins with a special focus on significant enrichment and/or depletion in each category.